Gene expression
Anna V. Protasio
01/11/2019
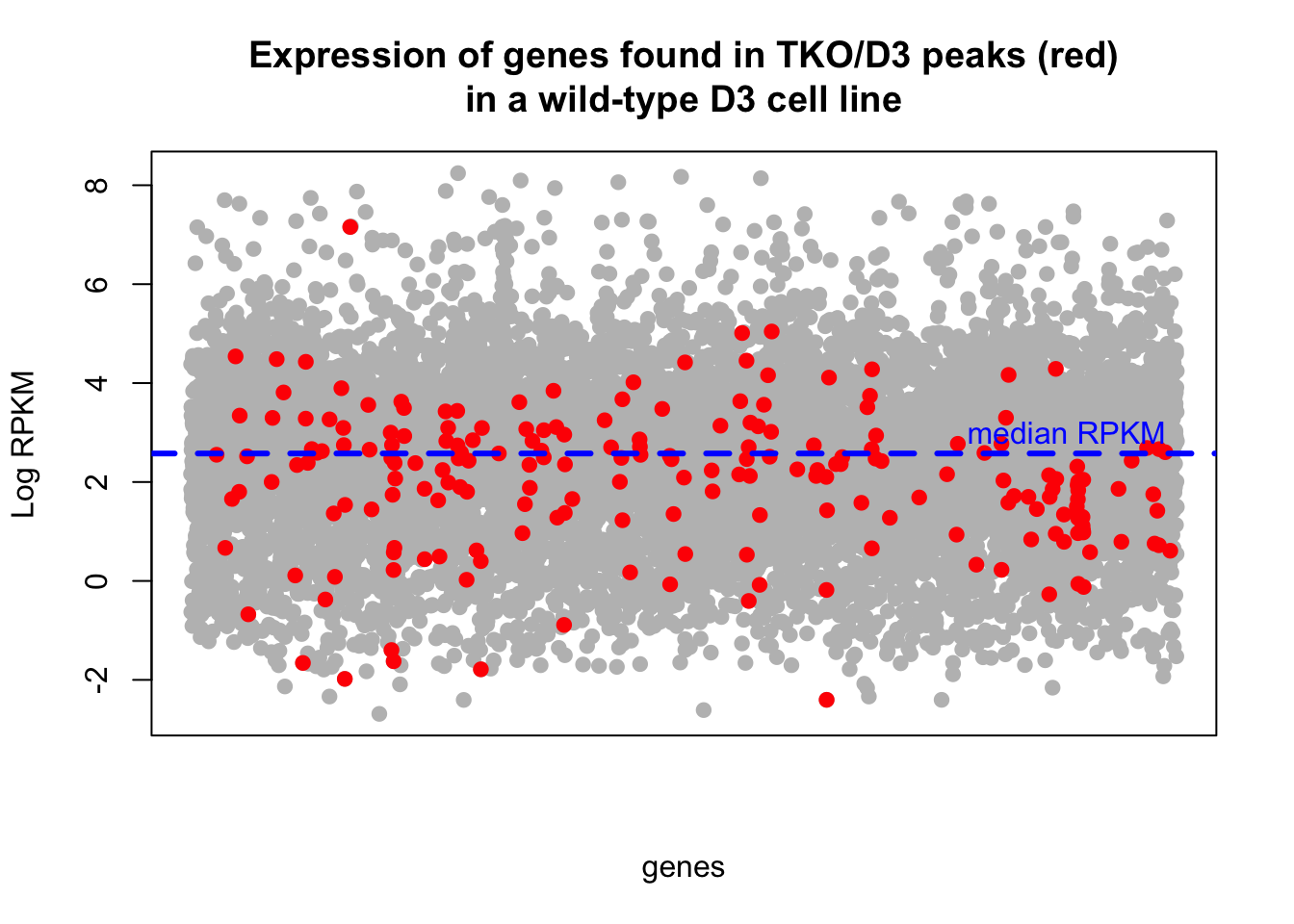
In this section we look at the expression of genes regulated by TKO/TASOR. We found these genes in previous steps by looking at the overlap between peaks that dissapear in TKO samples with respect to D3 and the gene annotation.
We then used RNAseq data from a “D3” sample, remove gene with less than min.count reads and computed RPKMs using the length of added exons (to account only for the coding region of the gene)
We can repeat the analysis with triplicates for the counts of D3 sample. In this case, the total read counts for each gene is calculated as the mean of the three samples obtained.
## R version 3.6.1 (2019-07-05)
## Platform: x86_64-apple-darwin15.6.0 (64-bit)
## Running under: macOS Mojave 10.14.6
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/3.6/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_GB.UTF-8/en_GB.UTF-8/en_GB.UTF-8/C/en_GB.UTF-8/en_GB.UTF-8
##
## attached base packages:
## [1] parallel stats4 stats graphics grDevices utils datasets
## [8] methods base
##
## other attached packages:
## [1] edgeR_3.26.8 limma_3.40.6 ggplot2_3.2.1
## [4] GenomicRanges_1.36.1 GenomeInfoDb_1.20.0 IRanges_2.18.3
## [7] S4Vectors_0.22.1 BiocGenerics_0.30.0 tidyr_1.0.0
## [10] kableExtra_1.1.0 RColorBrewer_1.1-2 knitr_1.25
##
## loaded via a namespace (and not attached):
## [1] locfit_1.5-9.1 tidyselect_0.2.5 xfun_0.10
## [4] purrr_0.3.2 lattice_0.20-38 colorspace_1.4-1
## [7] vctrs_0.2.0 htmltools_0.4.0 viridisLite_0.3.0
## [10] yaml_2.2.0 rlang_0.4.0 pillar_1.4.2
## [13] withr_2.1.2 glue_1.3.1 GenomeInfoDbData_1.2.1
## [16] lifecycle_0.1.0 stringr_1.4.0 zlibbioc_1.30.0
## [19] munsell_0.5.0 gtable_0.3.0 rvest_0.3.4
## [22] evaluate_0.14 Rcpp_1.0.2 readr_1.3.1
## [25] scales_1.0.0 backports_1.1.5 webshot_0.5.1
## [28] XVector_0.24.0 gridExtra_2.3 hms_0.5.1
## [31] digest_0.6.21 stringi_1.4.3 dplyr_0.8.3
## [34] grid_3.6.1 tools_3.6.1 bitops_1.0-6
## [37] magrittr_1.5 RCurl_1.95-4.12 lazyeval_0.2.2
## [40] tibble_2.1.3 crayon_1.3.4 pkgconfig_2.0.3
## [43] zeallot_0.1.0 xml2_1.2.2 viridis_0.5.1
## [46] assertthat_0.2.1 rmarkdown_1.16 httr_1.4.1
## [49] rstudioapi_0.10 R6_2.4.0 compiler_3.6.1